Mooney Lab @ University of Southern California
Welcome! We are the Mooney Lab. Our goal is to use patterns of variation in the genome to understand the evolutionary and population histories of both humans and other species. We do this by implementing and developing computational and statistical methods to study the genome.
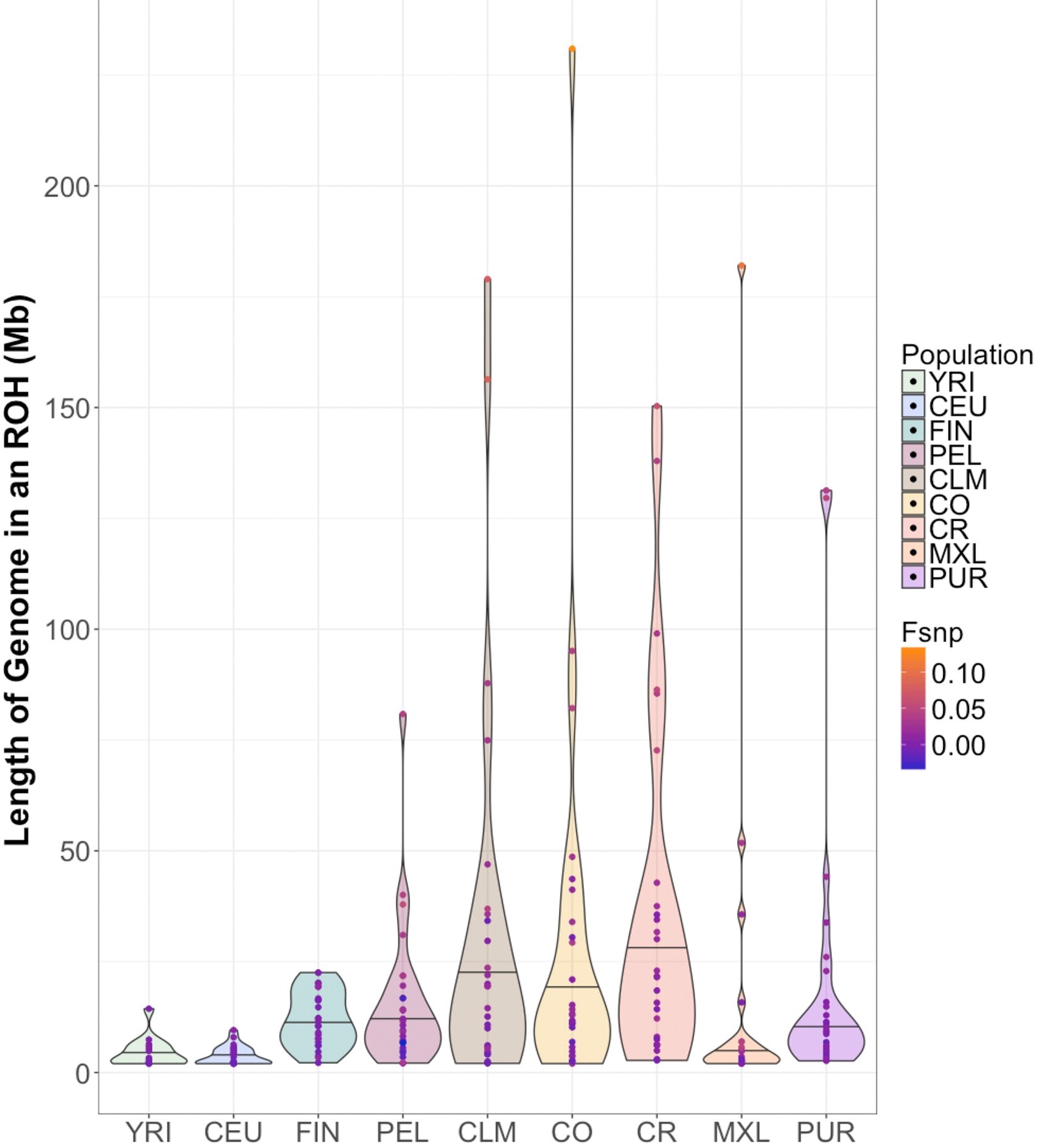
We are also interested in more broad population genetics questions such as: the genomic consequences of deleterious (non-neutral) mutations, where deleterious mutations tend to aggregate in the genome, and understanding patterns of genomic sharing through identity-by-descent segments and runs of homozygosity.

We are located at the University of Southern California (USC) in the Department of Quantitative and Computational Biology (QCB).
Our Research
For the last decade, population genetics has rapidly accelerated to new heights with the availability of sequence data. These data sets have provided valuable insights about population history, phenotypes, disease architecture, evolution, and genomic diversity.
Broadly, our lab uses empirical data analysis, simulation frameworks, and methods development to do the following: 1) infer demography, 2) understand the fitness impact of recessive variation, and 3) investigate complex trait architecture.
We focus primarily on integrating data from both human and non-human populations to develop a more complete picture of mechanisms that shape patterns of genomic sharing (in the form of identity-by-descent segments and runs of homozygosity) and deleterious variation. We are also interested in developing methods for conservation biology, inference of the genealogical histories of admixed populations, and the equitable inclusion of underrepresented populations in genomics.
Jazlyn's talk at CGSI
On-going research in the Mooney Lab on uncovering the history of African Americans.
Join the Mooney Lab
We are glad you are interested in joining us!
We are an inclusive lab. I support my students and believe they all deserve an oppurtunity to be in computational biology. The Mooney lab is a space where lab members not only learn the science but also are excited about the future of science. There are no required pre-requisites for joining the lab and lab members may come from any background. In the Mooney lab, most projects will include coding for genetic data analysis, some mathematics, simulation, and data visualization. Most of all, I want students to learn relevant skills for the next step in their career, in a lab where they feel like science is a place where they can grow and thrive.

Postdoctoral researchers
To inquire about postdoctoral positions, please write to Jazlyn at jazlynmo@usc.edu. Interest areas that would fit particularly well in the lab include the population genetics of runs of homozygosity, identity-by-descent segments, complex traits, and demographic inference.Please include a CV and a short description (1-2 pages) of your research experience, interests, and ideas for work in the lab. Please put “Postdoc position” in the subject line of your email to jazlynmo@usc.edu. Please also list 2-3 professional references and include any recent work you’d like to share.
PhD students
Feel free to contact me and put “Graduate student position” in the subject line of your email to jazlynmo@usc.edu. We can talk about potential research interests and relevant projects in the lab.Students should apply to USC’s Computational Biology and Bioinformatics (CBB) program. Our graduate program will give you experience in algorithms, statistics, and biology. During the first year, students will complete coursework as well as rotate in up to three CBB-affiliated labs. Students interested in completing a PhD in the lab should arrange to do one of their rotations with us. CBB admissions are handled by the admissions committee and not by individual labs.
Undergraduates, Master’s students, and others
We are looking to include USC undergraduates and Master's students on research projects. If you are interested in joining the lab, email me with the relevant header of either “Undergraduate student position” or " Master’s student position" in the subject line of your email to jazlynmo@usc.edu. Please include a CV, relevant course history, and statement of interest.Experience with command line, Python, R, C/C++, would be beneficial but are not necessarily required.
Contact Us
Email: jazlynmo@usc.edu
We are located on the 4th floor of Ray R. Irani Hall (RRI) at the University of Southern California, in the University Park neighborhood of Los Angeles.
The closest parking structure is the Downey Way Structure.
USC Maps
Our Publications
2025
Agranat-Tamir L, Agwamba KD, Mooney JA, & Rosenberg NA. Shared ancestors and the birthday problem. Am Stat. 2025 Dec 05:10.1080/00031305.2025.259597. doi:10.1080/00031305.2025.2595972.
Akopyan M, Genchev M, Armstrong EE, Mooney JA. Reference genome choice compromises population genetic analyses. Cell. 2025 Nov 26;188(24):6939-6952.e11. doi: 10.1016/j.cell.2025.08.034. Epub 2025 Sep 22. PMID: 40987293. PDF
Sherman CA&, Nataneli S&, Claw KG, Mooney JA. Echoes of eugenics: confronting its effects in indigenous genomics. Genetics. 2025 Aug 11:iyaf127. doi: 10.1093/genetics/iyaf127. Epub ahead of print. Erratum in: Genetics. 2025 Sep 18:iyaf172. doi: 10.1093/genetics/iyaf172. PMID: 40796125. & indicates co-first authorship
Ferrari T, Feng S, Zhang X+, Mooney J+. Parameter Scaling in Population Genetics Simulations May Introduce Unintended Background Selection: Considerations for Scaled Simulation Design. Genome Biol Evol. 2025 May 30;17(6):evaf097. doi: 10.1093/gbe/evaf097. PMID: 40405504; PMCID: PMC12147024. + indicates co-last authorship
2024
Kreger J, Mooney, JA, Shibata D, & MacLean AL. Developmental hematopoietic stem cell variation explains clonal hematopoiesis later in life. Nat Commun. 2024 Nov 26;15(1):10268. doi: 10.1038/s41467-024-54711-2. PMID: 39592593; PMCID: PMC11599844.
Armstrong EE&, Li C&, Campana MG, Ferrari T, Kelley JL, Petrov DA, Solari KA, Mooney, JA. A Pipeline and Recommendations for Population and Individual Diagnostic SNP Selection in Non‐Model Species. . Mol Ecol Resour. 2025 Apr;25(3):e14048. doi: 10.1111/1755-0998.14048. Epub 2024 Nov 29. PMID: 39611246; PMCID: PMC11887608. & indicates co-first authorship
Armstrong EE*,&, Mooney JA*,&, Solari KA, Kim BY, Barsh GS, Grant VB, Greenbaum G, Kaelin CB, Panchenko K, Pickrell JK, Rosenberg N, Ryder OA, Yokoyama T, Ramakrishnan U, Petrov DA, Hadly EA*. Unraveling the genomic diversity and admixture history of captive tigers in the United States. Proc Natl Acad Sci U S A. 2024 Sep 24;121(39):e2402924121. doi: 10.1073/pnas.2402924121. Epub 2024 Sep 19. PMID: 39298482; PMCID: PMC11441546. * indicates co-corresponding & indicates co-first authorship
Armstrong EE, Bissell KL, Fatima HS, Heikkinen MA, Jessup A, Junaid MO, Lee DH, Lieb EC, Liem JT, Martin EM, Moreno M, Otgonbayar K, Romans BW, Royar K, Adler MB, Needle DB, Harkess A, Kelley JL, Mooney JA* & Mychajliw, AM*. Chromosome-level assembly of the gray fox (Urocyon cinereoargenteus) confirms the basal loss of PRDM9 in Canidae. G3 (Bethesda). 2024 Apr 3;14(4):jkae034. doi: 10.1093/g3journal/jkae034. PMID: 38366575; PMCID: PMC10989890. * indicates co-corresponding
Agranat-Tamir L, Mooney JA, Rosenberg NA. Counting the genetic ancestors from source populations in members of an admixed population. Genetics. 2024 Apr 3;226(4):iyae011. doi: 10.1093/genetics/iyae011. PMID: 38289724; PMCID: PMC10990421.
2023
Mooney JA, Agranat-Tamir L, Pritchard JK, Rosenberg NA. On the number of genealogical ancestors tracing to the source groups of an admixed population. Genetics. 2023 Jul 6;224(3):iyad079. doi: 10.1093/genetics/iyad079. PMID: 37410594; PMCID: PMC10324943.
Mooney JA, Marsden CD, Yohannes A, Wayne RK, Lohmueller KE. Long-term small population size, deleterious variation, and altitude adaptation in the Ethiopian wolf, a severely endangered canid. Mol Biol Evol. 2023 Jan 4;40(1):msac277. doi: 10.1093/molbev/msac277. PMID: 36585842; PMCID: PMC9847632.
2021
Mooney JA, Yohannes A, Lohmueller KE. The impact of identity by descent on fitness and disease in dogs. Proc Natl Acad Sci U S A. 2021 Apr 20;118(16):e2019116118. doi: 10.1073/pnas.2019116118. PMID: 33853941; PMCID: PMC8072400.
2019
Sura SA, Smith LL, Ambrose MR, Amorim CEG, Beichman AC, Gomez ACR, Juhn M, Kandlikar GS, Miller JS, Mooney JA, Mummah RO, Lohmueller KE, Lloyd-Smith JO. Ten simple rules for giving an effective academic job talk. PLoS Comput Biol. 2019 Jul 25;15(7):e1007163. doi: 10.1371/journal.pcbi.1007163. PMID: 31344032; PMCID: PMC6657819.
2018
Mooney JA&, Huber CD&, Service S, Sul JH, Marsden CD, Zhang Z, Sabatti C, Ruiz-Linares A, Bedoy G, Costa Rica/Colombia Consortium for Genetic Investigation of Bipolar Endophenotypes, Freimer N, Lohmueller KE. Understanding the hidden complexity of Latin American population isolates.Am J Hum Genet. 2018 Nov 1;103(5):707-726. doi: 10.1016/j.ajhg.2018.09.013. Epub 2018 Oct 25. PMID: 30401458; PMCID: PMC6218714. & indicates co-first authorship
The Team

Jazlyn Mooney (she/her)
Principal Investigator (jazlynmo@usc.edu)Jazlyn is a Gabilan Assistant Professor in the Quantitative and Computational Biology (QCB) Department within the Dornsife College of Letters, Arts and Sciences at USC. Her research combines computational approaches with population genetics theory to better understand genetic variation, medical genetics, and human evolution. Jazlyn's CV
Jazlyn completed her undergraduate degree at the University of New Mexico where she studied human evolution with Jeffrey Long in the department of Anthropology. She completed her PhD in Human Genetics at UCLA in 2020 under the advisement of Kirk Lohmueller. At UCLA, Jazlyn studied genetic variation in admixed populations, complex traits in dogs, and conservation genomics. Afterward, she moved to Stanford’s Biology department for postdoctoral research with Noah Rosenberg; where she continued to study admixed populations with a focus on inference method development.
Fun fact from Jazlyn: When I am not in lab, I enjoy record collecting and going to concerts.

Shirin Nataneli (she/her/hers)
Graduate studentShirin is a graduate student in the Computational Biology and Bioinformatics program and is broadly interested population genetics and evolution. Currently, she is exploring how we can better model complex admixture scenarios to infer demography in humans and populations of conservation concern.
Fun fact from Shirin: I've collected snow globes from every place I've traveled and my favorite animal is a sheep.

Aydin Loid Karatas (he/him/his)
Graduate studentAydin is a graduate student in the Computational Biology and Bioinformatics program. Aydin is excited about the intersection of modeling human disease and large scale simulation frameworks to model them. He is currently exploring how admixture affects the distribution of identity by descent segments and eventually the deleterious variation within them.
Fun fact from Aydin: I own (and use) several film cameras and older digicams.

Shengmiao (Morgan) Huang (she/her/hers)
Graduate studentShengmiao is a graduate student in the Computational Biology and Bioinformatics program. She is interested in developing mathematical models for conservation biology. Currently, she is exploring how we can better quantify complex relatedness in small populations and developing software to infer relatedness in species of conservation concern.
Fun fact from Shengmiao: Water is my natural habitat. And I have the best cat in the world.

Tessa Ferrari (she/her/hers)
Incoming Graduate StudentTessa recently graduated from the Quantitative Biology undergraduate program had a short stint as our lab Research Technician. This fall she will be starting her PhD in the Computational Biology and Bioinformatics program. Tessa is interested in building and using large-scale simulation frameworks to infer demography and developing methods to quantify recessive mutational load while controlling for structure.
Fun fact from Tessa: In my free time I’m a competitive hip hop and open style dancer!

Matthew Genchev
Undergraduate studentMatthew is an undergraduate in the Quantitative Biology program with an interest in modeling recombination and demography in non-human populations. Matthew is currently exploring recombination and demography in Canidae, specifically foxes. Matthew was a Provost Undergraduate Research Fellow, and a Summer Undergraduate Research Fellow (SURF) . Now he is a Undergraduate Research Associates Program (URAP) Fellow .
Fun fact from Matthew: I won $20 from a scratch off ticket one time.

Sydney Bruce
Undergraduate studentSydney is an undergraduate in the Quantitative Biology program with an interest in population genetics and systems biology. Currently, Sydney is working to create a recombination map for captive tigers in the U.S. and will be exploring the relationship between genetic diversity and the recombination landscape in tigers. She is also a WiSE Undergraduate Fellow.
Fun fact from Sydney: I work as an EMT during the summers!

Tina Lasisi (she/her/hers)
Former Postdoctoral FellowDr. Tina Lasisi is now an Assistant Professor at the University of Michigan in the Anthropology department, and a former Postdoctoral Researcher in the Mooney and Edge labs. Tina's background and interests fall within the realm of Biological Anthropology and complex trait genetics. Specifically, her research area includes understanding the evolution of human variation in 1) pigmentation and 2) scalp hair. Tina is also active in science communication. Check out Tina's website and her amazing PBS mini-series Why Am I Like This?.
Fun fact from Tina: I have a standard poodle.

Maria Akopyan (she/her/hers)
Former Postdoctoral FellowDr. Maria Akopyan completed her PhD at Cornell where she studied gene flow and adaptation in Atlantic silversides. Dr. Akopyan holds an M.S., which she completed at CSUN, and studied variation in Red-eyed Tree frogs. While in our group, she was exploring how reference bias affects demographic inference in Canidae by using whole genome sequence data from Mainland foxes. She is a now an NSF PRFB postdoc in the Samuk and Armstrong labs at UCR.
Fun fact from Maria: I like to paint and dance!
Mengdi (Kim) Chai
Former Undergraduate studentMengdi is an alumni of the Mooney Lab and graduated with honors from the Quantitative Biology undergraduate program. While in the lab, Mengdi was a WiSE Undergraduate Fellow and used identity-by-descent segments to identify genes associated with coat color in tigers and tracking their ancestral origins. She recently completed her Master's Degree at Harvard in Computational Biology and Quantitative Genetics and will be working at Eli Lilly as a Marketing Management Trainee.
Fun fact from Kim: I have one hamster and he is cute! I decide to have two dogs, one cat, and two hamsters when I have my own apartment!

Anika Shrivastava
Former Undergraduate studentAnika was an undergraduate in the Quantitative Biology program and is interested in human genetics and disease. While in the lab, she worked on quantifying regions of the genome that harbor potential recessive lethal mutations by layering information from multiple types of genomic annotations. Anika was a Provost Undergraduate Research Fellow, completed an internship at Expedia, and now works as a Software Engineer at Capital One.
Fun fact from Anika: I have been playing the piano for 15 years and have a minor in musical studies at USC.

Chenyang (Julie) Li
Former Undergraduate studentJulie was an undergraduate in Applied and Computational Mathematics & the Quantitative Biology program with an interest in conservation and disease. She created the population assignment algorithm for mPCRselect, which allows users to construct informative marker panels in populations of conservation concern. She was a WiSE Undergraduate Fellow, graduated with honors and received the USC Discovery Scholar Distinction. Julie now at Emory in their Population Biology, Ecology, and Evolution PhD program.
Fun fact from Julie: I've never dyed my hair. My reddish/brown hair color is my natural hair color.

Mahija Mogalipuvvu
Former Undergraduate studentMahija was an undergraduate in the Quantitative Biology program and co-led our QBIO 490 section called Multi-Omic Data Analysis. While in the lab, she worked on a project which aimed to better understanding the allele frequency spectra of cancer associated variants. Mahija is continuing research and preparing to enter medical school.
Fun fact from Mahija: I love chocolate milk.

Peyton Hall
Former Undergraduate studentPeyton was an undergraduate in the Quantitative Biology program who was interested in pursuing medicine. While in the lab, she decided to branch out and continue our work on captive tigers. Peyton’s project layered information from multiple types of genomic annotations to explore whether there were differences in annotation distributions across ancestries. Peyton is now in medical school at Howard!
Fun fact from Peyton: I am volunteer birth doula.

Joseph Caluya
Former Undergraduate studentJoseph was an undergraduate in the Quantitative Biology program and is interested in cancer biology. While in the lab, Joseph worked on data from TCGA where he sought to understand share and population specific differential expression in variants associated with breast cancer. Joseph is taking some time off before his next phase and continuing research.
Fun fact from Joseph: In my free time I like to needle felt animals, write/read poetry, and try out different skincare products.
Classes Taught
HUMGEN 19: How Genetics has Contributed to Racial Injustice in America and the World
Fall 2020Fiat Lux seminar I co-taught with Dr. Nelson Freimer at UCLA. We used a mixture of readings, presentations, and discussions to navigate the history of Genetics and its ties to Eugenics. Syllabus
BIOL 870: Biology Colloquium
Fall 2020Seminar based course I taught at SFSU. In this course students’ scientific curiosity was be piqued as they heard and interacted with biologists from a range of subfields. Syllabus
QBIO 105: Introduction to Quantitative Biology Seminar
Spring 2023Seminar based course I co-taught with Dr. Remo Rohs at USC. This course is the introductory seminar for students taking the QBIO major. It is ideally taken as freshman but it can be taken after a student‘s transfer into the QBIO program. The instructors introduce the general field of Quantitative Biology, its definition and role within the Biological Sciences, and its relationship with Chemistry, Computer Science, Engineering, Mathematics, Medicine, and Physics. Syllabus
QBIO 475: Statistical and Evolutionary Genetics
Fall 2022, Fall 2024, and Fall 2024QBIO capstone course I co-teach with Dr. Doc Edge at USC. This is an upper-division course designed to introduce quantitative biologists to some central ideas in the mathematical modeling and statistical analysis of genetic variation. Topics from evolutionary genetics and medical genetics will be explored using a mix of math, simulation, and data analysis. Topics may include genetic drift, natural selection, mutation, migration, population structure, and study designs for learning about genotype-phenotype relationships, such as family-based studies, analyses of gene-expression variation, and genome-wide association studies. Syllabus

QBIO 115w: Ethics in Biology, Medicine, and Statistics
Spring 2024 & Spring 2025Dornsife General Education (GE) course I created and teach at USC. Biology, medicine, and statistics have each made a large impact on understanding life sciences and human disease. This impact has been bolstered over the last 20 years with the integration of information from all three fields and the advancement of genetics and computation. However, these fields are also deeply rooted in 19th century eugenics. Unfortunately, these ties percolated into foundational scientific data and were used to establish biased public policies with rise of eugenics movement. This course will critically reflect on the origins of each field through a scientific perspective and explore modern research practices that offer hope for an equitable future. We will use a mixture of readings, presentations, and discussions to navigate these topics. Syllabus
Outreach and Science Communication
UCLA Association for Multi-Ethnic Bioscientists Advancement (AMEBA)
Co-founderDuring graduate school I co-founded AMEBA algonside of Drs. Andrew Lopez, Jessica Ochoa and Christopher Robles. The goal of AMEBA is to create a community and support the academic development of underrepresented students across Biosciences. AMEBA has now grown to 40+ students spanning Life Sciences, Physical Sciences, and Graduate Programs in Biosciences.
Introduction to Tidyverse for Computational Biology
SACNAS WorkshopsAttended SACNAS in 2020 and 2022 to conduct workshops that cover the basic statistics and plotting using tidyverse for computational biology.
Presentation 2022
Ethics in Genetics and Genomics
Paradigm PodcastI was super excited to be interviewed by former QBIO student Wade Boohar for the Paradigm Podcast, a series published by Shift SC, a technology ethics club at USC. We got to explore many ethical considerations when dealing with population genetics and large-scale genomic studies.
Genetics, Ethics, and the Responsibility of Science
What are the origins of genetics and what are ethical considerations of large scale genomic studies?
Elephant in the Room: Small Talk, Big Issues
Natural History Museum of Los AngelesI got the opportunity to join a panel discussion about Dire Wolves for NHM’s new discussion series Elephant in the Room! This series is open to the public and explores bold–and sometimes controversial–topics from the museum world and beyond. Our panel, hosted by Charles Annenberg Weingarten, explored both established facts and emerging mysteries about dire wolves. With perspectives from paleontology and genomics, we addressed common misconceptions and discussed exciting developments in biotechnology, sequencing, and even de-extinction.
Dire Wolves: Still Extinct?
What do we know about Dire Wolves? Are they still extinct?
Lab in the news
Reference genome choice compromises population genetic analyses
News about our work on using non-species matched reference genomes led by former postdoc Dr. Maria Akopyan.Want to save an endangered species? Start with the right DNA blueprint
Unraveling the genomic diversity and admixture history of captive tigers in the United States
News about our work on Tigers with Dr. Ellie Armstrong.DNA Reveals the Origin Stories of America's Captive Tigers
The genetic secrets of the United States’s privately owned tigers
How captive tigers can help the wild ones: Q&A with USC’s Jazlyn Mooney
On the number of genealogical ancestors tracing to the source groups of an admixed population
News about our work on uncovering lost ancestors of African Americans.Mathematical model calculates probability of shared ancestors for African Americans
Study sheds light on Black Americans' ancestral links
A new approach to genetic genealogy sheds light on African American ancestry
New study offers African American genealogical information unrecoverable from written record
Researchers illuminate centuries of identity lost because of slavery
Breaking down the ‘brick wall’ of African American genealogy